SEABED
Plataforma web para apoyar en la investigación microbiológica del sector agrotech.
En colaboración con:
Pedro Palazón y Raquel Navarro, de Ideagro

Plataforma web para apoyar en la investigación microbiológica del sector agrotech.
En colaboración con:
Pedro Palazón y Raquel Navarro, de Ideagro

Inteligencia Artificial aplicada a la selección de verracos para mejorar el proceso de inseminación.
En colaboración con:
Dr. Francisco Alberto García, del departamento de Fisiología (Facultad de Veterinaria) de la Universidad de Murcia

Control de la bioestimulación microbiológica en cultivos.
En colaboración con:
Ramón Alcaraz, de Naturbec
Estudio de los efectos de una dieta antiinflamatoria en pacientes con artritis reumatoide y fibromialgia.
En colaboración con:
Dra. Lucrecia Moreno, Dr. Francisco J. Muñoz y Teresa López de la UCHCEU, Juan F. Martínez de ADM-Biopolis.
Identificación de parásitos intestinales en sardinas y alachas mediante técnicas metagenómicas.
En colaboración con:
Dra. María Auxiliadora Dea y Dra. Samantha Moratal del grupo de investigación en Salud Pública del UCH-CEU

Búsqueda de biomarcadores bacterianos en la microflora intestinal relacionados con enfermedades dérmicas.
En colaboración con:
Dr. Vicente Navarro y Laura Navarro de Bioithas.

Gracias a la ayuda de la App SpIA, serás capaz de clasificar de manera automática distintos tipos de células en imágenes de microscopía de dorada (Sparus aurata).
En colaboración con:
Dra. María Ángeles Esteban y María Ortiz del Departamento de Biología Celular e Histología de la Universidad de Murcia.
Análisis de metilación diferencial en ovocitos de vaca y cerdo madurados en distintas condiciones.
En colaboración con:
Dra. Pilar Coy y Dra. Laura Abril del Departamento de Fisiología de la Universidad de Murcia.

En la aplicación web ActinAItor se han integrado diferentes módulos de Inteligencia Artificial y análisis de imagen para facilitar la detección y el conteo de estructuras de actina.
En colaboración con:
Dr. José Cansado, Dra. María Isabel Madrid y Francisco Prieto, del Departamento de Genética y Microbiología de la Universidad de Murcia.

PredIActor es un software capaz de predecir la tasa de éxito en una inseminación artificial gracias al poder de la inteligencia artificial.
En colaboración con:
Dr. Manuel Avilés y la Dra. Paula Cots del Departamento de Biología Celular de la Universidad de Murcia.

Análisis de enriquecimiento e interpretación para la publicación científica "Effect of Superovulation Treatment on Oocyte’s DNA Methylation" de la revista International Journal of Molecular Sciences".
En colaboración con:
Dra. Pilar Coy del Departamento de Fisiología Veterinaria de la Universidad de Murcia.
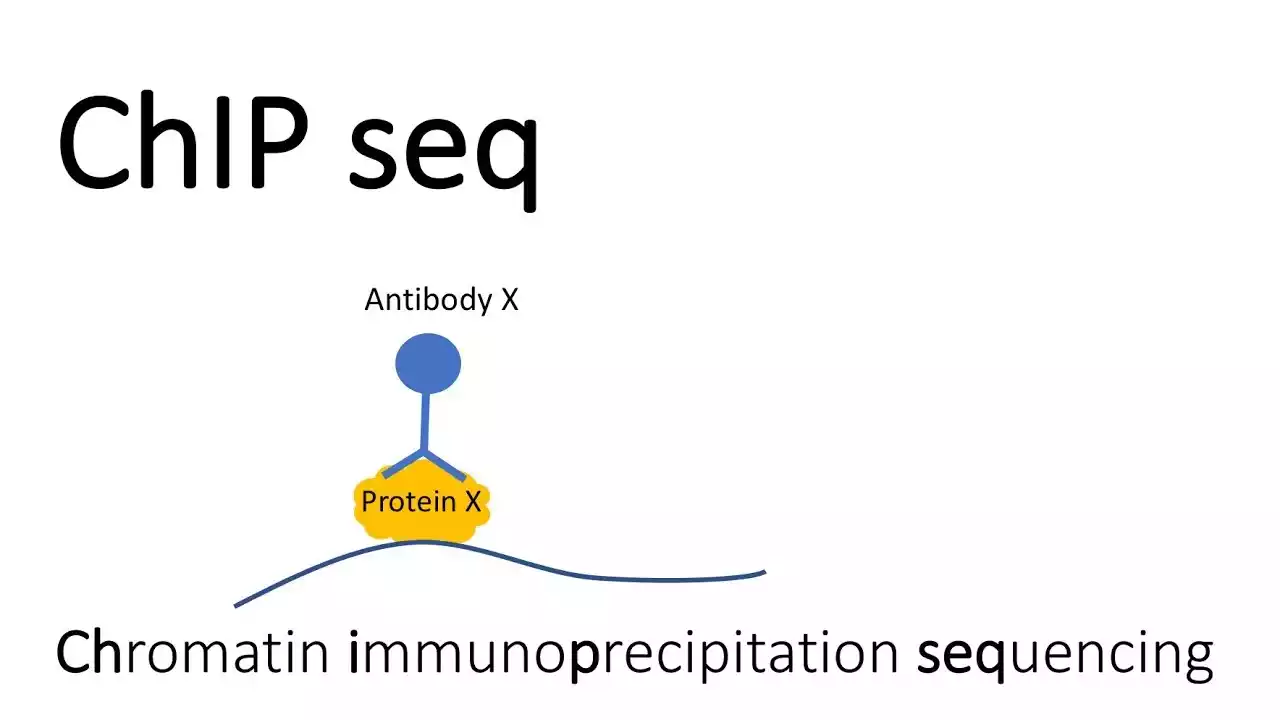
Análisis de zonas de unión de proteínas al ADN mediante la técnica ChIP-Seq en muestras de cerebro de Drosophila.
En colaboración con:
Dra. Alicia Hidalgo y Dra. María Dolores Pérez de la Universidad de Birmingham.

Análisis de datos de proteómica en doradas (Sparus aurata) para el estudio de la cicatrización.
En colaboración con:
Dra. María Ángeles Esteban y Dra. Nora Albaladejo, Dpto. Biología celular e histología de la Universidad de Murcia.

Alopecia areata (AA) is a chronic autoimmune disease characterized by non-scarring hair loss, where pathogenesis is closely linked to the collapse of hair follicle immune privilege and dysregulated T-cell responses. Increasing evidence suggests that gut dysbiosis may contribute to systemic immune alterations relevant to autoimmune disorders, yet its role in AA remains largely unexplored. In this study, we aimed to characterize the gut microbiota composition of AA patients and evaluate its potential as a biomarker for disease discrimination. Fecal samples from patients with AA and healthy controls were analyzed by 16S rRNA sequencing and processed through QIIME2 and MicrobiomeAnalyst platforms. Diversity metrics, differential abundance, and microbial network correlations were assessed, and supervised machine learning models were developed to classify AA versus control profiles. Our results revealed distinct microbial signatures in AA, with enrichment of pro-inflammatory genera such as Methanobrevibacter, Collinsella, and Ruminococcus gnavus, and depletion of immunoregulatory commensals, including Faecalibacterium and Eubacterium eligens group. Network analyses showed more complex microbial interactions in AA, and Random Forest models achieved 92% accuracy in discriminating AA from controls. These findings indicate that gut dysbiosis may play a role in AA pathogenesis, providing potential diagnostic biomarkers and supporting microbiota-targeted interventions as future therapeutic strategies.
Ver másÁngel Aguado-García,Francisco Huertas-López,David Martínez-Moreno,Emilio Manuel Serrano-López,María Martínez-Villaescusa,Carmen Carazo-Díaz & Vicente Navarro-López
Ver publicación
Guayule (Parthenium argentatum A. Gray) is a promising alternative for the sustainable production of natural rubber. In addition to rubber, guayule produces a wide range of secondary metabolites, such as resin, essential oils, and phenolic compounds, with a variety of potential applications in fields as diverse as biopesticides, biomedicine, cosmetics, and other industries. However, the dynamics of these metabolites in different accessions of guayule are not known in detail. Therefore, in the present study, a comprehensive multivariate analysis was performed with the data obtained by the research group over five years (82 metabolites in 27 accessions grown under standardized conditions) to evaluate their genetic and metabolic variability. Advanced statistical tools such as Principal Component Analysis (PCA), Hierarchical Cluster Analysis (HCA), and Spearman correlation were used. These analyses revealed that resin and rubber content alone are insufficient for accession differentiation. In contrast, sesquiterpenes, particularly guayulins, appeared to be strong biomarkers for genetic classification, correlating strongly with rubber and resin biosynthetic pathways. In addition, guayulin A and guayulin C seem to have a high correlation with rubber. With respect to guayulins, this analysis also seems to indicate that their biosynthesis occurs in the leaves, starting with guayulin C and D, and that they are then translocated to the stem. These results suggest that a comprehensive metabolite profiling approach, focusing on specific compound groups, can improve guayule breeding strategies aimed at maximizing rubber yield and resin utilization.
Ver másPedro J. García-Cano, M. Mercedes García-Martínez, Emilio J. González-Navarro, Amaya Zalacain, Grisel Ponciano, Dong Chen, Colleen McMahan, Manuel Carmona , Guayente Latorre
Ver publicación
Saline ponds are natural laboratories where microbial life adapts to intense osmotic stress. This year-long survey of ponds in San Pedro del Pinatar (Murcia, Spain) measured salinity, pH and temperature across a gradient and profiled microbial communities with 16S, 18S and ITS rRNA sequencing. Multivariate ordinations (PCoA, NMDS) showed salinity as the dominant structuring force. Low-salinity sites harboured diverse, mostly generalist bacteria and eukaryotes, whereas rising salinity pruned diversity and selected for specialists. Extreme ponds were overwhelmingly dominated by the haloarchaeon Haloquadratum walsbyi, the bacterium Salinibacter ruber, and halophilic fungi such as Hortaea and Wallemia. These extremophiles tolerate massive ionic imbalance through compatible-solute synthesis, specialised membrane lipids and DNA-repair systems. Their metabolic repertoire, including carotenoid antioxidants, salt-stable hydrolases and antimicrobial compounds, underscores the ponds’ biotechnological promise for green chemistry, food preservation, cosmetics and pharmaceutical lead discovery worldwide and tailored, salt-based processes for circular bioeconomy innovation. By linking physicochemical data to taxonomic turnover, our study maps ecological thresholds at which generalists yield to extreme specialists, emphasising hypersaline habitats as resilient yet vulnerable ecosystems. Future work should target genome-resolved metagenomics, cultivation of novel isolates and pilot bioprocesses, unlocking sustainable applications ranging from low-water bioremediation to enzymatic catalysis in high-salt industrial streams.
Ver másJuan Romero-Navarro, José Ramón Acosta-Motos, Santiago López-Miranda
Ver publicación
Many phenolic compounds, such as hydroxytyrosol (HT), have been recognized for their antioxidant and cardiovascular (CV) health benefits. To address the efficacy of HT, the present study aimed to identify the relevant mechanisms associated with high-CV risk. Plasma unbiased multi-omics data were compared among two subgroups of high-CV risk patients -HT responders and non-responders. The suppression of platelet reactivity and agonist-induced platelet activation observed after HT intervention were measured by CD61/CD62P expression and apoptotic microparticles with flow cytometry. Microbiota analysis revealed that HT treatment significantly increased and decreased the abundance of Ruminiclostridium sp. and Desulfovidrio sp., respectively. Lipid metabolism and proteomic responses were heterogeneous within the two distinct subgroups, associated mainly with thrombotic and hemostatic signals. The metabolomic analysis further confirmed the differentially expressed metabolites within these two subgroups, highlighting improved glutathione metabolism after HT treatment. The omics datasets integration offers a more comprehensive representation of the glutathione and coagulation pathways affected by the HT treatment. Even though the translation of multi-omics technologies into the clinical landscape is slow, the current interventional study provides an overview of their utility in developing novel therapeutic opportunities to prevent CV diseases, such as suppression of platelet reactivity.
Ver másClara Noguera-Navarro, Kasper T. Vinten, David Auñón-Calles, Carmen Carazo-Díaz, Georges E. Janssens and Silvia Montoro-García
Ver publicación
In vitro maturation (IVM) is a critical step in animal in vitro embryo production, yet oocytes matured in vitro often exhibit lower developmental competence than their in vivo counterparts. However, the molecular mechanisms behind this observation remain unclear. This study investigated the gene expression and DNA methylation profiles in porcine oocytes with different developmental competencies. To study these differences, we used as model oocytes from prepubertal gilts (IVM) and sows (in vivo matured) and assessed their developmental competence up to the blastocyst stage. We also examined their gene expression and DNA methylation profiles at single-cell resolution using RNA-sequencing and bisulfite-sequencing. Oocytes were obtained by aspiration of either ovarian follicles between 3–6mm diameter, and the subsequent IVM, or ovarian follicles from 8–10 mm diameter, with no need for maturation (in vivo matured oocytes). Cleavage rates (58.2 ± 3.0 and 45.7 ± 4.4) and blastocyst rates (31.4 ± 3.7 and 47.5 ± 6.6) for IVM and in vivo groups differed significantly. Using the in vivo group as a reference, IVM oocytes had 1297 down-regulated and 476 up-regulated differentially expressed genes (DEGs), with up-regulated DEGs associated with organelle organization and cell cycle processes, and down-regulated genes involved in protein synthesis and metabolomic processes. While global DNA methylation levels were similar between groups, a few differentially methylated regions were found in CpG islands, promoters, and coding regions. Our integrative analysis identified key methylated regions and genes that distinguish each group, suggesting that both donor age and maturation conditions significantly influence gene expression regulation in oocytes with different developmental competencies.
Ver másLaura Abril-Parreño, Jordana S Lopes, Jon Romero-Aguirregomezcorta, Antonio Galvao, Gavin Kelsey, Pilar Coy
Ver publicación
Diffuse large B-cell lymphoma (DLBCL) refers to an aggressive lymphoma that arises from germinal center (GC) B-cells, which differentiate into plasma cells (PC) to produce high affinity antibodies. 40% of DLBCL patients relapse or are refractory to the conventional immunochemotherapy treatment, usually with fatal consequences. Therefore, there is an unmet critical need to find more targeted therapies for DLBCL. DLBCL are characterized by profound alterations in the epigenome, which are correlated with poor survival. While epigenetic therapies are used as anti-cancer treatments, their full potential has not been achieved, mainly because of their limited efficacy when used as monotherapies and recurrent side effects associated with their low specificity. The abnormal epigenetic landscape of DLBCL tumors is associated with a blockade in GC exit and differentiation programs, which are regulated by the transcription factor BCL6. This aberrant repression of BCL6-target genes is mediated at least by two epigenetic mechanisms: 1) increased DNA methylation and 2) loss of acetylation of the lysine 27 of histone 3 (H3K27ac)-through recruitment of histone deacetylase 3 (HDAC3). Therefore, we investigated the efficacy against DLBCL of a novel combinatorial epigenetic therapy using the hypomethylating agent (HMA) 5-Azacitidine (5-Aza) and a specific HDAC3 inhibitor (HDAC3i). We found that treatment with 5-Aza and HDAC3i had a potent synergistic anti-tumor activity in vitro and in vivo, which was superior to the effect of each single drug or 5-Aza combined with non-specific HDACi and, importantly, was not associated with toxicity in normal cells. We also demonstrated that the combined 5-Aza and HDAC3i treatment induced the epigenetic remodeling of DLBCL cells, which resulted in a more potent re-expression of PC differentiation genes, including XBP1 and ATF4, compared to each drug used as single agents. Our results highlight the importance of targeting multiple layers of the epigenome to maximize the efficacy of epigenetic-based therapies.
Ver másCalixte Calaud, Emilio Serrano-Lopez, Leonie A. Cluse, Izabela Todorovski, Thomas Mardivirin, Matt Teater, Abdelillah Maarad, Noura Tawfic, Sophia Rutaquio, Peter J. Fraser, Killian Guiraud, David Yoannidis, Emeline Kerreneur, Arnaud Jacquel, Ari M. Melnick, Ricky W. Johnstone, Pilar M. Dominguez
Ver publicación
Background/Objectives: Growing evidence highlights the pivotal role of gut dysbiosis in the pathophysiology of irritable bowel syndrome (IBS). Despite this, the identification of an “IBS microbiota signature” remains elusive, primarily due to the influence of genetic, dietary, and environmental factors. To address these confounding variables, it is critical to perform comparative analyses using a control group derived from the same community as the IBS patients. This study aimed to evaluate and contrast the gut microbiota composition of IBS patients with healthy controls. Methods: We compared the gut microbiota from stool samples of 25 IBS patients diagnosed according to the Rome IV criteria, and 110 healthy subjects without acute or chronic diseases and not on continuous medication. The high-throughput sequencing of the V3–V4 regions of the 16S rRNA gene was conducted for microbiota analysis. Results: The IBS gut microbiota was richer but exhibited lower alpha diversity compared to the control group, suggesting simplification and imbalance. A beta diversity analysis revealed overall compositional differences between the two groups. A heat tree analysis highlighted key IBS-associated changes, including a decrease in Firmicutes, mainly due to Clostridia, and an increase in Bacteroidota, driven by an expansion of Bacteroidales families. Differential expression analyses identified important genera within these taxa like Bacteroides, Faecalibacterium, and Blautia, which could serve as microbiota-based biomarkers for IBS. Conclusions: Our results reveal both statistically and clinically significant differences in gut microbiota composition and diversity between IBS patients and healthy controls from the same community. These findings provide a deeper understanding of how alterations in the gut microbiota may contribute to IBS symptoms, offering new insights into the diagnosis and potential treatments.
Ver másPedro Sánchez-Pellicer, José María Álamo-Marzo, María Martínez-Villaescusa, Eva Núñez-Delegido, José Francisco Such-Ronda, Francisco Huertas-López, Emilio Manuel Serrano-López, David Martínez-Moreno and Vicente Navarro-López
Ver publicación
Endothelial injury, inflammatory infiltrate and fibrosis are the predominant lesions in the testis of bulls with besnoitiosis that may result in sterility. Moreover, fibroblasts, which are key players in fibrosis, are parasite target cells in a Besnoitia besnoiti chronic infection. This study aimed to decipher the molecular basis that underlies a drift toward fibrosis during the disease progression. Transcriptomic analysis was developed at two times post-infection (p.i.), representative of invasion (12 h p.i.) and intracellular proliferation (32 h p.i.), in primary bovine aorta fibroblasts infected with B. besnoiti tachyzoites. Once the enriched host pathways were identified, we studied the expression of selected differentially expressed genes (DEGs) in the scrotal skin of sterile infected bulls. Functional enrichment analyses of DEGs revealed shared hallmarks of cancer and early fibrosis. Biomarkers of inflammation, angiogenesis, cancer, and MAPK signaling stood out at 12 h p.i. At 32 h p.i., again MAPK and cancer pathways were enriched together with the PI3K–AKT pathway related to cell proliferation. Some DEGs were also regulated in the skin samples of naturally infected bulls (PLAUR, TGFβ1, FOSB). We have identified potential biomarkers and host pathways regulated during fibrosis that may hold prognostic significance and could emerge as potential therapeutic targets.
Ver másMaría Fernández-Álvarez, Pilar Horcajo , Alejandro Jiménez-Meléndez, Pablo Angulo Lara, Ana Huertas-López, Francisco Huertas-López, Ignacio Ferre, Luis Miguel Ortega-Mora and Gema Álvarez-García
Ver publicación
En la era de la medicina personalizada, las tecnologías de secuenciación y las herramientas ómicas han revoucionado la comprensión de las enfermedades, destacando la importancia de la microbiota intestinal en la salud y la enfermedad. Este estudio se centra en la relación entre la microbiota intestinal y dos enfermedades cutáneas inflamatorias: dermatitis atópica y alopecia areata. El objetivo es profundizar en estas conexiones a través de estudios bioinformáticos que permitan desarrollar un modelo de inteligencia artificial para su clasificación basada en icroorganismos específicos.
Ver másSánchez Pellicer P, Núñez Delegido E, Navarro Moratalla L, Agüera Santos JG, Serrano López EM, Huertas López F, Navarro López V
Ver publicación
Fish has core relevance in humans' diets all over the world, both for its importance in food security and for its enormous nutritional benefits. However, fish consumption is not exempt from risks, such as the presence of chemical pollutants or disease-causing biological agents, such as parasites. In particular, marine fish serve as hosts to a wide variety of parasites, including foodborne emerging zoonotic parasites, as nematodes of the genus Anisakis, of significant public health concern. Furthermore, they can also harbor other emerging zoonotic parasites originating from terrestrial environments, capable of reaching and contaminating marine waters. In both cases, the presence of these parasites in marine fish intended for human consumption represents a potential risk of transmission to humans. Given the distinct nature of these two groups of parasites and their varying research requirements, this doctoral thesis is structured into two parts.
Ver másSamantha Moratal Martínez, María Auxiliadora Dea Ayuela, Jordi López Ramon
Ver publicación
A lo largo del tracto reproductor femenino, el espermatozoide sufre una serie de cambios necesarios para que la fecundación tenga lugar, conocidos como capacitación. Uno de ellos es la hiperactivación, caracterizada por un cambio en el movimiento espermático necesario para que el espermatozoide pueda atravesar la zona pelúcida del óvulo. La hiperactivación se puede determinar de un modo no invasivo mediante el sistema automático CASA (Computer Assisted Sperm Analysis). El patrón de motilidad de los espermatozoides es heterogéneo y complejo, por ello el uso de modelos de inteligencia artificial (IA) resultan útiles a la hora de identificar parámetros relacionados con la hiperactivación, permitiendo el desarrollo de modelos computacionales que ayuden a la predicción de la calidad seminal y su potencial aplicación en la predicción del éxito en la inseminación artificial.
Ver másPaula Cots Rodríguez, T Casas Pina, EM Serrano López, M Avilés González, F Huertas López, J Gálvez Pradillo, J Álvarez Castillo, M Avilés
Ver publicación
Con la incorporación de las técnicas de reproducción asistida para tratar el problema de la infertilidad humana, surge el reto de combatir la pérdida de embarazo temprano. El embrión juega un papel clave en la eficiencia de la implantación que, unido a la necesidad de reducir el número de embriones a transferir, conduce por un lado a la mejora de las condiciones de cultivo y desarrollo embrionario, y por otro, al avance de las técnicas de selección embrionaria. Con este último propósito, el objetivo principal de esta tesis ha sido la identificación de polimorfismos (SNPs) propios del embrión relacionados con la infertilidad, susceptibles de convertirse en biomarcadores genéticos. Posteriormente, los resultados obtenidos fueron integrados en un sistema de Inteligencia Artificial (IA), junto con otros parámetros derivados del ciclo reproductivo de los pacientes, con el objeto de crear una herramienta de selección embrionaria capaz de predecir el éxito de la implantación y el desarrollo embrionario temprano.
Ver másNuria Hernández Terrés, Manuel Avilés Sánchez
Ver publicación
Controlled ovarian stimulation is a necessary step in some assisted reproductive procedures allowing a higher collection of female gametes. However, consequences of this stimulation for the gamete or the offspring have been shown in several mammals. Most studies used comparisons between oocytes from different donors, which may contribute to different responses. In this work, we use the bovine model in which each animal serves as its own control. DNA methylation profiles were obtained by single-cell whole-genome bisulfite sequencing of oocytes from pre-ovulatory unstimulated follicles compared to oocytes from stimulated follicles. Results show that the global percentage of methylation was similar between groups, but the percentage of methylation was lower for non-stimulated oocytes in the imprinted genes APEG3, MEG3, and MEG9 and higher in TSSC4 when compared to stimulated oocytes. Differences were also found in CGI of imprinted genes: higher methylation was found among non-stimulated oocytes in MEST (PEG1), IGF2R, GNAS (SCG6), KvDMR1 ICR UMD, and IGF2. In another region around IGF2, the methylation percentage was lower for non-stimulated oocytes when compared to stimulated oocytes. Data drawn from this study might help to understand the molecular reasons for the appearance of certain syndromes in assisted reproductive technologies-derived offspring.
Ver másJordana S. Lopes, Elena Ivanova, Salvador Ruiz, Simon Andrews, Gavin Kelsey, and Pilar Coy.
Ver publicación
Protein kinase C (PKC) comprises a family of highly related serine/threonine protein kinases involved in multiple signaling pathways, which control cell proliferation, survival, and differentiation. The role of PKCα in cancer has been studied for many years. However, it has been impossible to establish whether PKCα acts as an oncogene or a tumor suppressor. Here, we analyzed the importance of PKCα in cellular processes such as proliferation, migration, or apoptosis by inhibiting its gene expression in a luminal A breast cancer cell line (MCF-7). Differential expression analysis and phospho-kinase arrays of PKCα-KD vs. PKCα-WT MCF-7 cells identified an essential set of proteins and oncogenic kinases of the JAK/STAT and PI3K/AKT pathways that were down-regulated, whereas IGF1R, ERK1/2, and p53 were up-regulated. In addition, unexpected genes related to the interferon pathway appeared down-regulated, while PLC, ERBB4, or PDGFA displayed up-regulated. The integration of this information clearly showed us the usefulness of inhibiting a multifunctional kinase-like PKCα in the first step to control the tumor phenotype. Then allowing us to design a possible selection of specific inhibitors for the unexpected up-regulated pathways to further provide a second step of treatment to inhibit the proliferation and migration of MCF-7 cells. The results of this study suggest that PKCα plays an oncogenic role in this type of breast cancer model. In addition, it reveals the signaling mode of PKCα at both gene expression and kinase activation. In this way, a wide range of proteins can implement a new strategy to fine-tune the control of crucial functions in these cells and pave the way for designing targeted cancer therapies.
Ver másEmilio M Serrano-López, Teresa Coronado-Parra, Consuelo Marín-Vicente, Zoltan Szallasi, Victoria Gómez-Abellán, María José López-Andreo, Marcos Gragera, Juan C Gómez-Fernández, Rubén López-Nicolás, Senena Corbalán-García.
Ver publicación
Parkinson’s disease (PD) is a disabling neurodegenerative disorder in which multiple cell types, including dopaminergic and cholinergic neurons, are affected. The mechanisms of neurodegeneration in PD are not fully understood, limiting the development of therapies directed at disease-relevant molecular targets. C. elegans is a genetically tractable model system that can be used to disentangle disease mechanisms in complex diseases such as PD. Such mechanisms can be studied combining high-throughput molecular profiling technologies such as transcriptomics and metabolomics. However, the integrative analysis of multi-omics data in order to unravel disease mechanisms is a challenging task without advanced bioinformatics training. Galaxy, a widely-used resource for enabling bioinformatics analysis by the broad scientific community, has poor representation of multi-omics integration pipelines. We present the integrative analysis of gene expression and metabolite levels of a C. elegans PD model using GAIT-GM, a new Galaxy tool for multi-omics data analysis. Using GAIT-GM, we discovered an association between branched-chain amino acid metabolism and cholinergic neurons in the C. elegans PD model. An independent follow-up experiment uncovered cholinergic neurodegeneration in the C. elegans model that is consistent with cholinergic cell loss observed in PD. GAIT-GM is an easy to use Galaxy-based tool for generating novel testable hypotheses of disease mechanisms involving gene-metabolite relationships.
Ver másLauren M. McIntyre, Francisco Huertas, Alison M. Morse, Rachel Kaletsky, Coleen T. Murphy, Vrinda Kalia, Gary W. Miller, Olexander Moskalenko, Ana Conesa & Danielle E. Mor.
Ver publicación
The assessment of the safety of nano-biomedical products for patients is an essential prerequisite for their market authorization. However, it is also required to ensure the safety of the workers who may be unintentionally exposed to the nano-biomaterials (NBMs) in these medical applications during their synthesis, formulation into products and end-of-life processing and also of the medical professionals (e.g., nurses, doctors, dentists) using the products for treating patients. There is only a handful of workplace risk assessments focussing on NBMs used in medical applications. Our goal is to contribute to increasing the knowledge in this area by assessing the occupational risks of magnetite (Fe3O4) nanoparticles coated with PLGA-b-PEG-COOH used as contrast agent in magnetic resonance imaging (MRI) by applying the software-based Decision Support System (DSS) which was developed in the EU H2020 project BIORIMA.
Ver másV. Cazzagon, E. Giubilato, L. Pizzol, C. Ravagli, S. Doumett, G. Baldi, M. Blosi, A. Brunelli, C. Fito, F. Huertas, A. Marcomini, E. Semenzin, A. Zabeo, I. Zanoni, D. Hristozov.
Ver publicación
PKCε is highly expressed in mast cells and plays a fundamental role in the antigen-triggered activation of the allergic reaction. Although its regulation by diacylglycerols has been described, its regulation by acidic phospholipids and how this regulation leads to the control of downstream vesicle secretion is barely known. Here, we used structural and evolutionary studies to find the molecular mechanism that explains the selectivity of the C1B domain of PKCε by Phosphatidic Acid (PA). This resided in a collection of Arg residues that form a specific rim on the outer surface of the C1B domain, around the diacylglycerol binding cleft. In RBL-2H3 cells, this basic rim allowed the kinase to respond specifically to phosphatidic acid signals that induced its translocation to the plasma membrane and subsequent activation. Further experiments in cells that overexpress PKCε and a mutant of the PA binding site, showed that PA-dependent PKCε activation increased vesicle degranulation in RBL-2H3 cells, and this correlated with increased SNAP23 phosphorylation. Over-expression of PKCε in these cells also induced an increase in the number of docked vesicles containing SNAP23, when stimulated with PA. This accumulation could be attributed to the stabilizing effect of phosphorylation on the formation of the SNARE complex, which ultimately led to increased release of content in the presence of Ca2+ during the fusion process. Therefore, these findings reinforce the importance of PA signaling in the activation of PKCε, which could be an important target to inhibit the exacerbated responses of these cells in the allergic reaction.
Ver másEmilio M. Serrano-López, David López-Martínez, Juan C. Gómez-Fernández, Antonio Luis Egea-Jiménez, Senena Corbalán-García.
Ver publicación
During the last decade, the use of nanomaterials, due to their multiple utilities, has exponentially increased. Nanomaterials have unique properties such as a larger specific surface area and surface activity, which may result in health and environmental hazards different from those demonstrated by the same materials in bulk form. Besides, due to their small size, they can easily penetrate through the environmental and biological barriers. In terms of exposure potential, the vast majority of studies are focused on workplace areas, where inhalation is the most common route of exposure. The main route of entry into the environment is due to indirect emissions of nanomaterials from industrial settings, as well as uncontrollable releases into the environment during the use, recycling and disposal of nano-enabled products. Accidental spills during production or later transport of nanomaterials and release from wear and tear of materials containing nanomaterials may lead to potential exposure. In this sense, a proper understanding of all significant risks due to the exposure to nanomaterials that might result in a liability claim has been proved to be necessary. In this paper, the utility of an application for smartphones developed for the insurance sector has been validated as a solution for the analysis and evaluation of the emerging risk of the application of nanotechnology in the market. Different exposure scenarios for nanomaterials have been simulated with this application. The results obtained have been compared with real scenarios, corroborating that the use of novel tools can be used by companies that offer risk management in the form of insurance contracts.
Ver másFrancisco Aznar Mollá,Carlos Fito-López, Jose Antonio Heredia Alvaro and Francisco Huertas-López.
Ver publicación
Killer-cell immunoglobulin-like receptors (KIR) are expressed by natural killer (NK) and effector T cells. Although KIR+ T cells accumulate in oncologic patients, their role in cancer immune response remains elusive. This study explored the role of KIR+CD8+ T cells in cancer immunosurveillance by analyzing their frequency at diagnosis in the blood of 249 patients (80 melanomas, 80 bladder cancers, and 89 ovarian cancers), their relationship with overall survival (OS) of patients, and their gene expression profiles. KIR2DL1+ CD8+ T cells expanded in the presence of HLA-C2-ligands in patients who survived, but it did not in patients who died. In contrast, presence of HLA-C1-ligands was associated with dose-dependent expansions of KIR2DL2/S2+ CD8+ T cells and with shorter OS. KIR interactions with their specific ligands profoundly impacted CD8+ T cell expression profiles, involving multiple signaling pathways, effector functions, the secretome, and consequently, the cellular microenvironment, which could impact their cancer immunosurveillance capacities. KIR2DL1/S1+ CD8+ T cells showed a gene expression signature related to efficient tumor immunosurveillance, whereas KIR2DL2/L3/S2+CD8+ T cells showed transcriptomic profiles related to suppressive anti-tumor responses. These results could be the basis for the discovery of new therapeutic targets so that the outcome of patients with cancer can be improved.
Ver másLourdes Gimeno, Emilio M Serrano-López, José A Campillo, María A Cánovas-Zapata, Omar S Acuña, Francisco García-Cózar, María V Martínez-Sánchez, María D Martínez-Hernández, María F Soto-Ramírez, Pedro López-Cubillana, Jorge Martínez-Escribano, Jerónimo Martínez-García, Senena Corbalan-García, María R Álvarez-López, Alfredo Minguela.
Ver publicación
In recent years, nanomaterials have gained tremendous attention due to their wide variety of industrial applications including food packaging, consumer products, nanomedicines, etc. The fascinating properties of nanoparticles which are responsible for creating several exciting opportunities, however, are also accountable for growing concerns of their toxic effects on humans as well as the environment. Thus, in the present study, the authors have developed generalized models for predicting the cytotoxicity and genotoxicity of seven metal oxide nanoparticles. The models not only take into account the structural features, but also the diverse experimental conditions under which the toxicity of nanoparticles was determined. The diverse experimental conditions were captured in the generalized models using the Box-Jenkins moving average approach. Here, two machine learning techniques, namely, linear discriminant analysis and random forest were utilized to build the final models. Importantly, the validation metrics showed that the developed models have significant discriminatory power.
Ver másPravin Ambure, Arantxa Ballesteros, Francisco Huertas, Pau Camilleri, Stephen J. Barigye, Rafael Gozalbes.
Ver publicación
Altered methylation patterns are driving forces in colorectal carcinogenesis. The serrated adenocarcinoma (SAC) and sporadic colorectal carcinoma showing histological and molecular features of microsatellite instability (hmMSI-H) are two endpoints of the so-called serrated pathological route sharing some characteristics but displaying a totally different immune response and clinical outcome. However, there are no studies comparing the methylome of these two subtypes of colorectal carcinomas. The methylation status of 450,000 CpG sites using the Infinium Human Methylation 450 BeadChip array was investigated in 48 colorectal specimens, including 39 SACs and 9 matched hmMSI-H. Microarray data comparing SAC and hmMSI-H showed an enrichment in functions related to morphogenesis, neurogenesis, cytoskeleton, metabolism, vesicle transport and immune response and also significant differential methylation of 1540 genes, including CD14 and HLA-DOA which were more methylated in hmMSI-H than in SAC and subsequently validated at the CpG, mRNA and protein level using pyrosequencing, quantitative polymerase chain reaction (qPCR) and immunohistochemistry. These results demonstrate particular epigenetic regulation patterns in SAC which may help to define key molecules responsible for the characteristic weak immune response of SAC and identify potential targets for treating SAC, which lacks molecular targeted therapy.
Ver másJosé García-Solano, María C. Turpin, Daniel Torres-Moreno, Francisco Huertas-López, Anne Tuomisto, Markus J. Mäkinen, Ana Conesa & Pablo Conesa-Zamora.
Ver publicación
The intrinsic characteristics of the tumor microenvironment (TME), including acidic pH and overexpression of hydrolytic enzymes, offer an exciting opportunity for the rational design of TME-drug delivery systems (DDS). We developed and characterized a pH-responsive biodegradable poly-L-glutamic acid (PGA)-based combination conjugate family with the aim of optimizing anticancer effects. We obtained combination conjugates bearing Doxorubicin (Dox) and aminoglutethimide (AGM) with two Dox loadings and two different hydrazone pH-sensitive linkers that promote the specific release of Dox from the polymeric backbone within the TME. Low Dox loading coupled with a short hydrazone linker yielded optimal effects on primary tumor growth, lung metastasis (∼90% reduction), and toxicological profile in a preclinical metastatic triple-negative breast cancer (TNBC) murine model. The use of transcriptomic analysis helped us to identify the molecular mechanisms responsible for such results including a differential immunomodulation and cell death pathways among the conjugates. This data highlights the advantages of targeting the TME, the therapeutic value of polymer-based combination approaches, and the utility of –omics-based analysis to accelerate anticancer DDS.
Ver másJuan J. Arroyo-Crespo, Ana Armiñán, David Charbonnier, Leandro Balzano-Nogueira, Francisco Huertas-López, Cristina Martí, Sonia Tarazona, Jerónimo Forteza, Ana Conesa, María J. Vicent.
Ver publicación